NₓClinical Software
NₓClinical is an industry-leading decision support software solution for the detection of genome-wide variation from NGS and array data.
Comprehensive and up-to-date decision support solution for cytogenomics and molecular genetics to assist with the analysis and interpretation of genomic variants from microarray and NGS data.
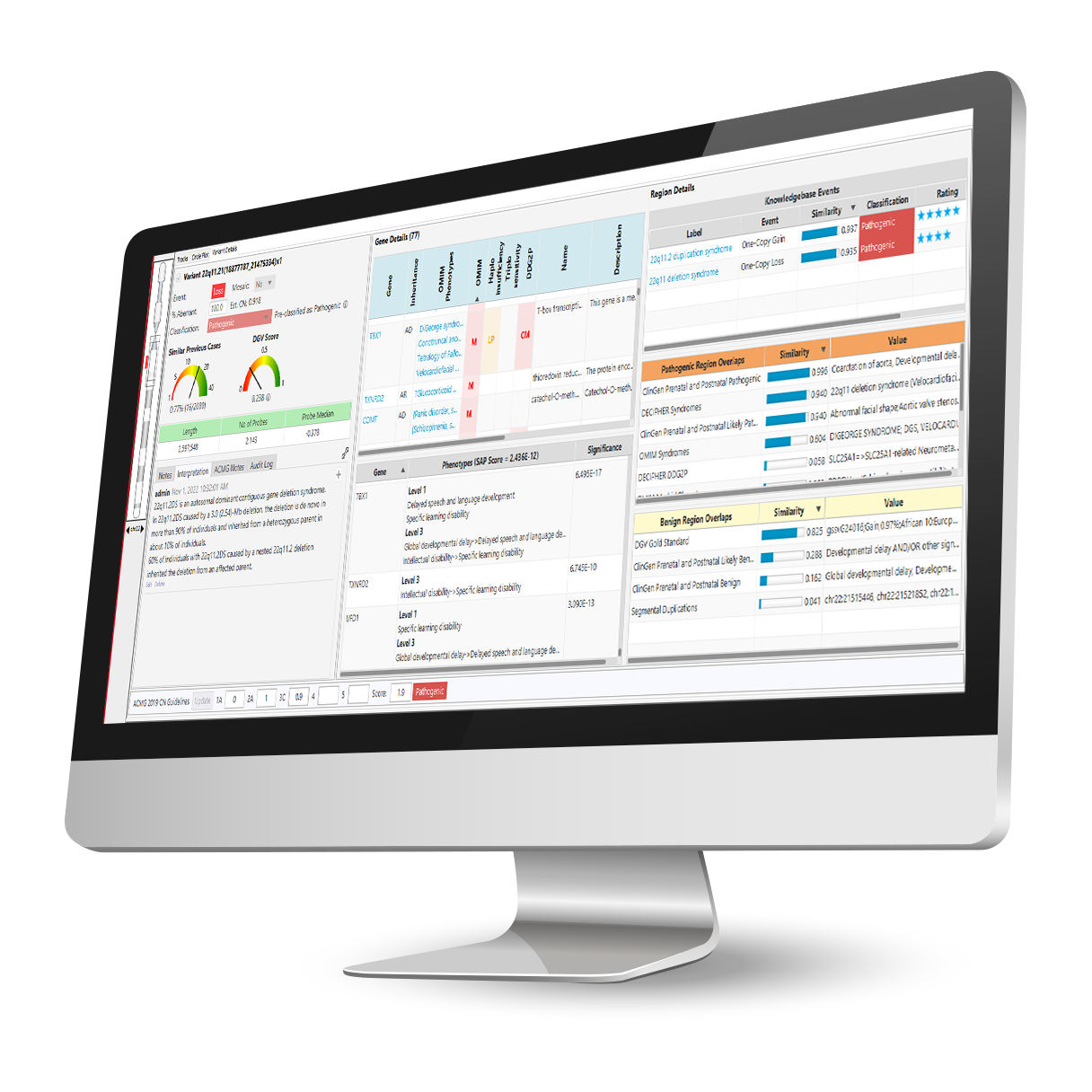
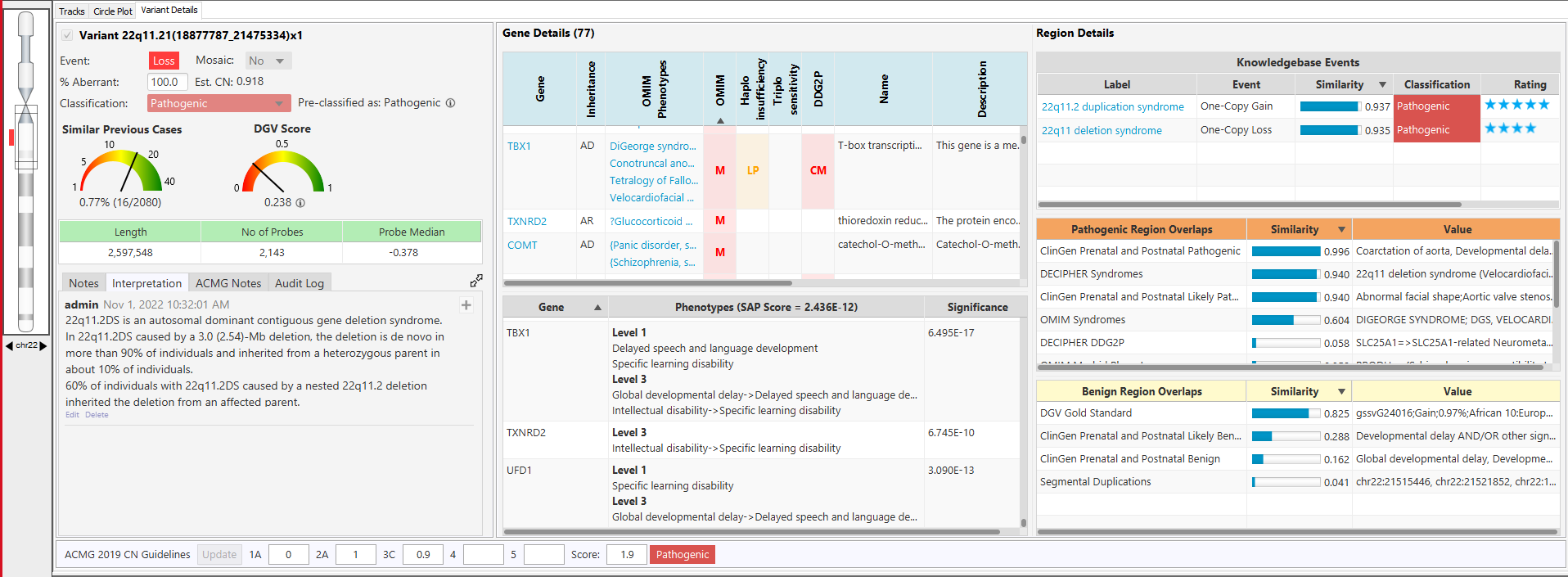
With the NₓClinical software, you get:
NₓClinical offers multi-platform integration by:
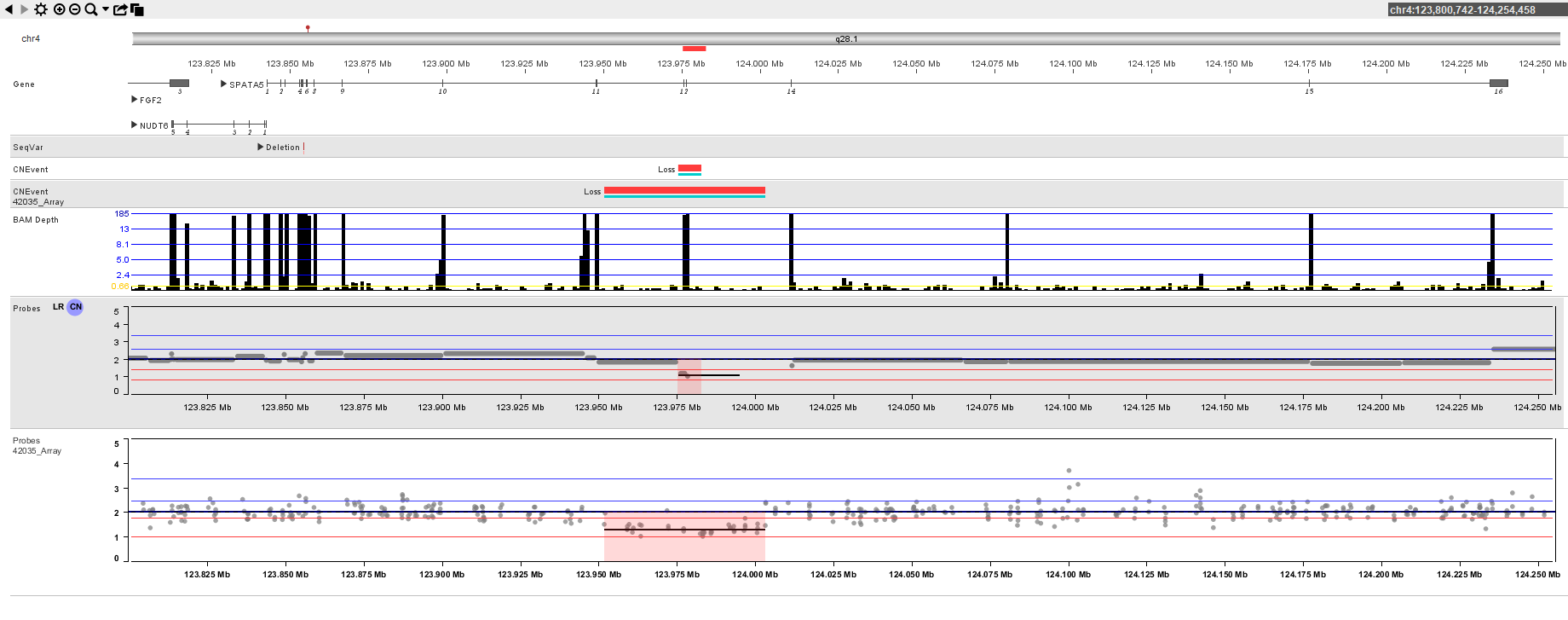

Bionano is committed to providing:
See how PerkinElmer Genomics uses the power of NₓClinical’s capabilities to call CNVs from NGS.
View WebinarRead more about homologous recombination deficiency and how Bionano NxClinical software provides in-depth analysis of HRD.
Read White PaperLearn how Ambry Genetics used NxClinical software to increase their case review efficiency by 5X.
Review Case StudySchedule a time to review the software to see how it can detect and analyze genome-wide variations.
Request a Software Demo